In case you have to convince someone on which horse to bet:
Tuesday, December 18, 2012
Lenovo x220 Trackpoint and Ubuntu
Although I'm using my Lenovo x220 laptop for more than a year, I discovered the settings for the track-point yesterday (random searches and forums, don't remember). I was never really thinking about it, but the track-point was a bit slow and stiff. The configuration settings of the usual UI configuration menu had no effect at all (yes, certainly a bug), either.
So, to reveal the magic, here it is. I've added a udev configuration file here:
$ cat /etc/udev/rules.d/trackpoint.rules
SUBSYSTEM=="serio", DRIVERS=="psmouse", WAIT_FOR="/sys/devices/platform/i8042/serio1/serio2/sensitivity", ATTR{sensitivity}="240", ATTR{speed}="95", ATTR{press_to_select}="1", ATTR{inertia}="6"
This is just one line for the settings of sensitivity, speed and intertia. You can also play around with them manually, e.g.
$ echo "5" | sudo tee /sys/devices/platform/i8042/serio1/serio2/inertia
So, to reveal the magic, here it is. I've added a udev configuration file here:
$ cat /etc/udev/rules.d/trackpoint.rules
SUBSYSTEM=="serio", DRIVERS=="psmouse", WAIT_FOR="/sys/devices/platform/i8042/serio1/serio2/sensitivity", ATTR{sensitivity}="240", ATTR{speed}="95", ATTR{press_to_select}="1", ATTR{inertia}="6"
This is just one line for the settings of sensitivity, speed and intertia. You can also play around with them manually, e.g.
$ echo "5" | sudo tee /sys/devices/platform/i8042/serio1/serio2/inertia
Friday, November 30, 2012
Mathematica 9's R integration vs. Rpy2
Some notes about "What's new in Mathematica 9: Builtin R Integration"
To understand what's going on, please open the example: Hierarchical Clustering.
This posting is basically about how to integrate R via Rpy2 in Python and hence also in Sage.
First thing you should notice is the clash of two parallel worlds. MMA's "RSet" command converts and sets the variable y in the scope of R. That's nice, but wouldn't it be much easier, to just have a local variable for that? Second of all, the function definitions inside strings - filled with escaped quotes - is also not that great. Isn't there are better quoting available?
What I want to do is to accomplish something similar with Rpy2. So yes, there are also equivalents to this RSet function, and you can also evaluate arbitrary R code inside strings (Python has triple-quoting to avoid escaping quotes). But that's not everything. You can import R packages [from rpy2.robjects.packages import importr] and bind them to local variables and have, for example, tab-completion on them. R Functions can be referenced directly, data-sets can be exchanged with implicit converters (e.g. numpy's ndarray -> R's Matrix), etc.
You can see the final output and code here: https://gist.github.com/4176508
Besides the usual import/from stuff common in Python, I start by creating a random matrix with Numpy:
mdata = np.random.randn(10, 5)
Enable autmatic conversion between Numpy and R:
from rpy2.robjects import numpy2ri
numpy2ri.activate()
Print the matrix using R's print:
rprint = robj.globalenv.get("print")
rprint(mdata)
[,1] [,2] [,3] [,4] [,5]
[1,] 2.1844065 -1.05401295 1.18316261 -0.356338229 1.55031790
[2,] 1.2476473 0.89507075 -0.14576584 0.006899727 0.01350773
[3,] -1.6416017 -0.28180113 0.02784612 1.199042583 0.01272994
[4,] 1.8645490 0.30993270 0.18107913 0.131505590 0.57083588
[5,] 1.2397427 0.73639680 -0.42883124 -0.436741492 0.43644592
[6,] -0.1980955 -0.04463804 -0.11217381 1.768415923 -1.82884840
[7,] 0.2943145 2.12648235 0.21068166 1.718289719 0.15711455
[8,] 0.2453526 0.64922040 1.80518277 0.086208024 1.18789962
[9,] 0.8229507 0.56227084 -1.72153433 -1.511514201 1.04610492
[10,] 0.1430900 0.03371198 -0.58992825 1.023002088 -1.47153121
... and just for fun, R'summary, printed with Python's print function:
print r.summary(mdata)
V1 V2 V3 V4
Min. :-1.6416 Min. :-1.05401 Min. :-1.72153 Min. :-1.5115
1st Qu.: 0.1687 1st Qu.:-0.02505 1st Qu.:-0.35806 1st Qu.:-0.2655
Median : 0.5586 Median : 0.43610 Median :-0.04216 Median : 0.1089
Mean : 0.6202 Mean : 0.39326 Mean : 0.04097 Mean : 0.3629
3rd Qu.: 1.2457 3rd Qu.: 0.71460 3rd Qu.: 0.20328 3rd Qu.: 1.1550
Max. : 2.1844 Max. : 2.12648 Max. : 1.80518 Max. : 1.7684
V5
Min. :-1.82885
1st Qu.: 0.01292
Median : 0.29678
Mean : 0.16746
3rd Qu.: 0.92729
Max. : 1.55032
... now applying the labels. This time, I show how to execute R code directly, hence injecting the variable "y" in R's global namespace, too:
from rpy2 import robjects as robj
robj.globalenv['y'] = mdata
r("""
dimnames(y) <- b="b">list(paste("g", 1:10, sep=""),
paste("t", 1:5, sep=""))
y
""")
Compare this to the way you have to do this in MMA9!
--- EDIT
Below is a way how the same is accomplished in Python. The paste command is replaced by Python's list comprehension, the list command is from R and mdata is converted to an R object to be able to do slot assignments.
mdata = numpy2ri.numpy2ri(mdata)
import rpy2.rinterface as ri
descr = ri.baseenv["list"](
ri.StrSexpVector(['g%s'%_ for _ in range(10)]),
ri.StrSexpVector(['t%s'%_ for _ in range(5)]))
mdata.do_slot_assign("dimnames", descr)
The first example is certainly more pleaseant, but once you would create some neat aliases for the R functions (e.g. list = ri.baseenv["list"]; strv = ri.StrSexpVector; ...), working in Python shouldn't be hard, too.
--- END EDIT
Calculating the correlation and distance matrix. Notice, that dots in R's functions, like the as.dist(), are converted to underscores in Python. Those dots are just like normal letters in the identifier name, nothing further.
stats = importr("stats")
corrm = r.cor(r.t(mdata), method="spearman")
# no idea how to do 1-matrix automagically
robj.globalenv['corrm'] = corrm
distm = stats.as_dist(r("1-corrm"))
rprint(corrm)
rprint(distm)
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
[1,] 1.0 -0.3 0.1 0.5 -0.2 0.9 0.8 0.6 0.3 0.2
[2,] -0.3 1.0 0.6 0.3 0.7 -0.4 0.0 0.1 0.2 -0.5
[3,] 0.1 0.6 1.0 -0.1 -0.1 -0.3 -0.1 0.7 0.6 -0.9
[4,] 0.5 0.3 -0.1 1.0 0.7 0.6 0.7 -0.1 -0.3 0.3
[5,] -0.2 0.7 -0.1 0.7 1.0 0.0 0.3 -0.4 -0.3 0.2
[6,] 0.9 -0.4 -0.3 0.6 0.0 1.0 0.9 0.3 0.1 0.6
[7,] 0.8 0.0 -0.1 0.7 0.3 0.9 1.0 0.4 0.3 0.5
[8,] 0.6 0.1 0.7 -0.1 -0.4 0.3 0.4 1.0 0.9 -0.4
[9,] 0.3 0.2 0.6 -0.3 -0.3 0.1 0.3 0.9 1.0 -0.3
[10,] 0.2 -0.5 -0.9 0.3 0.2 0.6 0.5 -0.4 -0.3 1.0
1 2 3 4 5 6 7 8 9
2 1.3
3 0.9 0.4
4 0.5 0.7 1.1
5 1.2 0.3 1.1 0.3
6 0.1 1.4 1.3 0.4 1.0
7 0.2 1.0 1.1 0.3 0.7 0.1
8 0.4 0.9 0.3 1.1 1.4 0.7 0.6
9 0.7 0.8 0.4 1.3 1.3 0.9 0.7 0.1
10 0.8 1.5 1.9 0.7 0.8 0.4 0.5 1.4 1.3
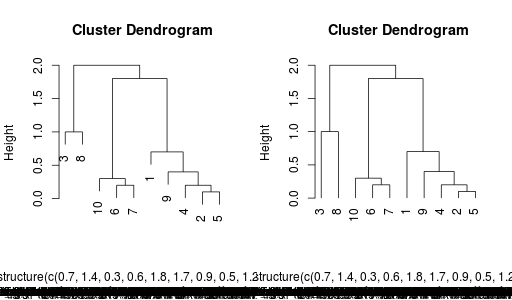
The clustering happens below (note: I had to execute the "NULL" in the r context, because it seems that Python's "None" isn't converted)
hr = stats.hclust(distm, method = "complete", members = r("NULL"))
Plotting is straightforward too. To plot to a device besides X11, one has to be a bit more specific. The documentation is full of examples. Notice, the only special part is the mfrow=r.c(1,2).
Looking at Wolfram's blogpost, I really don't want to understand what MMA's "getRPlot[...]" calling "mathematicaRPlotWrapper" does.
grdevices = importr('grDevices')
grdevices.png(file="mma9rpy2.png", width=512, height=300)
try:
r.par(mfrow = r.c(1,2))
r.plot(hr, hang = 0.1)
r.plot(hr, hang = -0.1)
finally:
grdevices.dev_off()
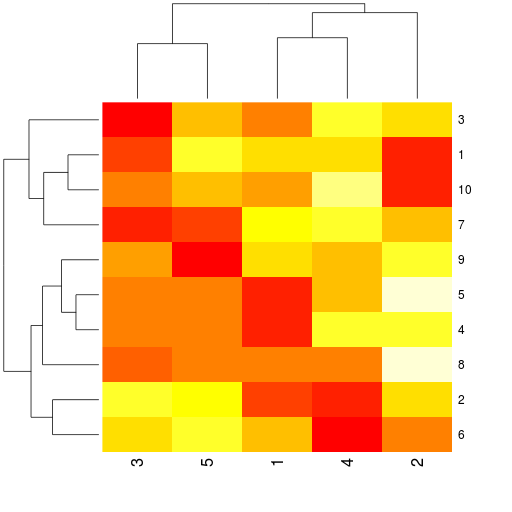
grdevices.png(file="mma9rpy2-2.png", width = 512, height = 512)
try:
r.heatmap(mdata)
finally:
grdevices.dev_off()
Final results:
... and a heatmap plot:
Final note, yes I know there is some fuzz at the bottom of the image with the dendrogram. I don't know why. Maybe someone can fix this ... and yes, that's possible, because all of this is fully open-sourced :-)
td;dr: https://gist.github.com/4176508
Friday, November 23, 2012
Sage 5.4.1 Released
Sage 5.4.1 was released on 15 November 2012.
It is available in source and binary form from:
Sage (http://www.sagemath.org/) is developed by volunteers and combines over 90 open source packages. For instructions about installing Sage, see
The following page lists the platforms on which Sage should work:
If you have any questions and/or problems, please report them to any of these Google groups:
The following 15 people contributed to this release. Of those, 2 made their first contribution to Sage:
- Aly Deines
- Benjamin Hutz [first contribution]
- Burcin Erocal
- David Loeffler
- Dmitrii Pasechnik
- Jeroen Demeyer
- John Palmieri
- Karl-Dieter Crisman
- Kenneth Smith
- Paul Zimmermann
- Punarbasu Purkayastha
- Sarah Chisholm
- Sebastien Gouezel [first contribution]
- Travis Scrimshaw
- Volker Braun
* Release manager: Jeroen Demeyer.
* We closed 13 tickets in this release. For details, see
http://boxen.math.washington.
Closed tickets:
#13309: Build Sage on OS X Mountain Lion [Reviewed by Dmitrii Pasechnik]
Merged in sage-5.4.1.rc0:
#6367: Karl-Dieter Crisman, Kenneth Smith: polygon2d -- several issues: typo in docs, shouldn't have been renamed [Reviewed by Volker Braun]
#10803: Paul Zimmermann: critical bug in real_roots [Reviewed by Jeroen Demeyer]
#12753: Benjamin Hutz: is_PrimeField import error [Reviewed by David Loeffler]
#12859: Aly Deines: quaternion algebra 'ramified at one prime' [Reviewed by Sarah Chisholm]
#13382: Dmitrii Pasechnik: build docs for SymmetricGroupRepresentation(
It is available in source and binary form from:
Sage (http://www.sagemath.org/) is developed by volunteers and combines over 90 open source packages. For instructions about installing Sage, see
The following page lists the platforms on which Sage should work:
If you have any questions and/or problems, please report them to any of these Google groups:
- sage-support: http://groups.google.com/
group/sage-support - sage-devel: http://groups.google.com/
group/sage-devel
The following 15 people contributed to this release. Of those, 2 made their first contribution to Sage:
- Aly Deines
- Benjamin Hutz [first contribution]
- Burcin Erocal
- David Loeffler
- Dmitrii Pasechnik
- Jeroen Demeyer
- John Palmieri
- Karl-Dieter Crisman
- Kenneth Smith
- Paul Zimmermann
- Punarbasu Purkayastha
- Sarah Chisholm
- Sebastien Gouezel [first contribution]
- Travis Scrimshaw
- Volker Braun
* Release manager: Jeroen Demeyer.
* We closed 13 tickets in this release. For details, see
http://boxen.math.washington.
Closed tickets:
#13309: Build Sage on OS X Mountain Lion [Reviewed by Dmitrii Pasechnik]
Merged in sage-5.4.1.rc0:
#6367: Karl-Dieter Crisman, Kenneth Smith: polygon2d -- several issues: typo in docs, shouldn't have been renamed [Reviewed by Volker Braun]
#10803: Paul Zimmermann: critical bug in real_roots [Reviewed by Jeroen Demeyer]
#12753: Benjamin Hutz: is_PrimeField import error [Reviewed by David Loeffler]
#12859: Aly Deines: quaternion algebra 'ramified at one prime' [Reviewed by Sarah Chisholm]
#13382: Dmitrii Pasechnik: build docs for SymmetricGroupRepresentation(
#13533: Jeroen Demeyer: Remove "optional - gcc" from doctests [Reviewed
by Karl-Dieter Crisman, John Palmieri]
#13541: John Palmieri: update scipy to 0.11.0 [Reviewed by Dmitrii Pasechnik]
#13598: John Palmieri: 'x' should be defined when using 'sage -c' [Reviewed by Punarbasu Purkayastha]
#13632: Sebastien Gouezel: Fix latex display of arguments of symbolic functions [Reviewed by Burcin Erocal]
Merged in sage-5.4.1.rc1:
#13407: Jeroen Demeyer: Move sage-make_relative to sage-location [Reviewed by Dmitrii Pasechnik]
#13452: Jeroen Demeyer: Refactor sage-location [Reviewed by Dmitrii Pasechnik]
#13689: Jeroen Demeyer: Fix upgrading from relocated Sage with GCC [Reviewed by John Palmieri]
by Karl-Dieter Crisman, John Palmieri]
#13541: John Palmieri: update scipy to 0.11.0 [Reviewed by Dmitrii Pasechnik]
#13598: John Palmieri: 'x' should be defined when using 'sage -c' [Reviewed by Punarbasu Purkayastha]
#13632: Sebastien Gouezel: Fix latex display of arguments of symbolic functions [Reviewed by Burcin Erocal]
Merged in sage-5.4.1.rc1:
#13407: Jeroen Demeyer: Move sage-make_relative to sage-location [Reviewed by Dmitrii Pasechnik]
#13452: Jeroen Demeyer: Refactor sage-location [Reviewed by Dmitrii Pasechnik]
#13689: Jeroen Demeyer: Fix upgrading from relocated Sage with GCC [Reviewed by John Palmieri]
Thursday, November 15, 2012
Sage 5.4 released
I'm glad to blog (and "reshare) that Sage 5.4 is available now.
Some random picks from the release notes:
- notebook internationalization
- 4ti2 interface
- MathJax instead of jsMath
- Bijection between Rigged Configurations and Crystal Paths
- new features in group algebra category
- non commutative symmetric functions
- Update Cremona's table of elliptic curves to 270000
- Plancherel measure of an individual partition
... and much more besides even more bugfixes :-)
http://sagemath.org/download.html
And I suggest you to follow Sage at Google+ to stay in the loop.
New Mirror: Universidade de São Paulo
Sage just got a second mirror in South America, at the Universidade de São Paulo: http://linorg.usp.br/sage. Now, there are two in Brazil and no one in another country, but still better than just one SPOF.
Monday, April 30, 2012
Sage goes GSoC
This year Sage is part of the GSoC program for the first year. This is an exciting new opportunity to get new contributors on board and to aim for new features. We got nearly 60 project proposals, of which about 8 or more were really excellent ones. We got 3 slots to fill and therefore it was a really hard decision which projects to pick. We are curious how this will turn out during the summer.
The projects are:
The projects are:
- Lattices by Jan Pöschko: Jan will work on a new "Lattice" class for Sage.
- Univariate polynomials and factorization library by Lina Kulakova: here, the aim is to implement a factorization for polynomials over F_p where p is large.
- Symbolic Expressions by Titus Nicolae: this project aims to improve Sage's core code for handling symbolic expressions.
The actual work starts on May 21st and all three students will give regular updates through their blogs or by posting to the mailing list to keep others in the loop what they are doing. All regular developers are invited to give them feedback and help them if they encounter any problems.
Subscribe to:
Posts (Atom)